Reprogramming, stem cells and oncogenesis

Objectives
Our laboratory is made up of developmental, reprogramming, cancer biologists and bioinformaticians. We elucidate the mechanisms that control embryonic development, and in particular the establishment of the cellular identities that make up our body. We also study how cell identity and plasticity are regulated during reprogramming and oncogenic transformation.
In the context of the Impulscience program from the Fondation Bettencourt Schueller, we propose fundings for passionate students, engineers and postdoctoral researchers. Send your spontaneous applications to the following address: Fabrice.lavial@lyon.unicancer.fr.

Cellular identity and plasticity are key parameters of embryonic development, cellular reprogramming and oncogenesis. Lineage differentiation is accompanied by a restriction of plasticity and the acquisition of specialized identities. On the contrary, a loss of identity and a regain of plasticity are observed when the epigenome of a cell is dedifferentiated and reprogrammed towards the pluripotent state into iPS cells (induced pluripotent stem cells) by the pioneer transcription factors Oct4, Sox2 , Klf4 and c-Myc (OSKM). Interestingly, epigenetic alterations of cell identity and plasticity are also emerging as crucial regulators of different stages of tumorigenesis in different adult and also pediatric contexts.
We address the following fundamental questions:
-What are the molecular mechanisms that control the establishment of cellular identities during development and how are they regulated during reprogramming and transformation? Our work will have an impact on reproductive biology, regenerative medicine and cancer biology.
-What are the trajectories used by a somatic cell when it changes identity and reprograms? Can this trajectory make the somatic cell inclined to acquire transformed properties?
-Can we reprogram the epigenome of a cancer cell or one undergoing transformation?
Axis 1: Control of cellular identity during reprogramming and oncogenic transformation
The team’s first line of research aims to dissect the early molecular events of cellular reprogramming and transformation. To do this, we carry out multi-omic analyzes at the single cell scale by combining the use of 2D cellular models, 3D organoids and murine genetic models. We recently published a proof-of-concept study demonstrating that cellular identity and cellular plasticity can be uncoupled and is controlled by dedicated gene regulatory networks (Huyghe A., Furlan G. et al., Nature Cell Biology 2022) (Huyghe A. et al., Trends in Cell Biology 2023)(Puisieux A. et al., Cancer Cell 2018). We attempt to identify cellular dynamics as well as the mechanisms promoting or limiting the change in the identity of a cell.
Lung organoid (B. Martin/A. Huyghe).
Axis 2: Pluripotent stem cell dynamics
The second research theme aims to understand the molecular mechanisms controlling the self-renewal of pluripotent stem cells, as well as their ability to polarize and self-assemble. Here, we use models of embryonic or induced stem cells, murine and human, cultured in 2D or induced to form 3D epiblastoid structures or synthetic embryos. We pioneered the stem cell field few years ago by identifying the ligand Netrin-1, previously known as an axon guidance cue, as a crucial regulator of naive pluripotency (Huyghe A., Furlan G., Ozmadenci D. et al., Nature Cell Biology 2020)(Furlan G. et al., Nature Communications 2023). We are now trying to understand how pluripotent stem cells integrate the external signals they receive, whether chemical or mechanical, and why and how they respond in an heterogeneous manner.
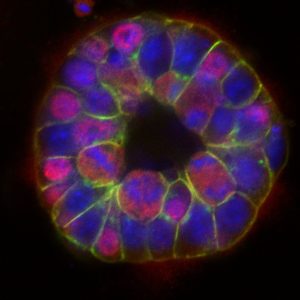
Epiblast-like structure mimicking an embryo generated by the self-assembly of pluripotent stem cells in hydrogel (M. Lantelme/E. Cascales).
-
Fabrice Lavial
Team leader